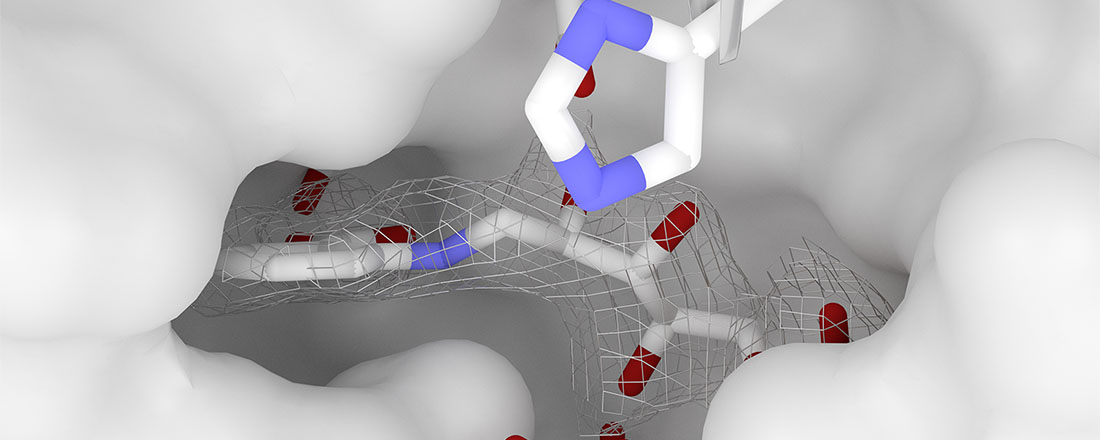
We study the evolution of protein folds and functions and apply this knowledge to protein design. Evolutionary mechanisms as observed in Nature can teach us about concepts to use in design, while design provides a way to test evolutionary hypotheses. In our studies we combine theoretical and experimental approaches.
The evolution of the frequently encountered (βα)8-barrel enzyme and its relationship to flavodoxin-like proteins is used to investigate how the contemporary protein folds evolved. Many (βα)8-barrels are believed to have evolved from a precursor half its size. Database searches revealed striking structural similarities between half-barrels and proteins adopting the flavodoxin-like fold. We built new proteins from fragments of these different folds thereby reconstructing possible evolutionary events. Todays enzymes are highly optimized. For many reactions the underlying mechanisms are not well understood, which is why their design is such a challenging task. By studying reactions in their evolutionary context we want to contribute to a more thorough understanding of protein function. We further are developing computational design tools such as ScaffoldSelection, which identifies suitable protein scaffolds for a given catalytic motif, and PocketOptimizer, which calculates changes in ligand binding pockets.